


New insights into microbial evolution
Photos by Evan Krape and courtesy of Terry Papoutsakis and John Hill | Photo illustration by Joy Smoker January 12, 2024
UD engineers uncover new mechanism for gene transfer with implications for fields ranging from ecology to biotechnology and medicine
If you still remember that “Dear King Phillip Came Over For Good Spaghetti,” you’ll likely also recall the corresponding taxonomic ranks of biology: Domain, Kingdom, Phylum, Class, Order, Family, Genus and Species. The two domains include prokaryotes, single-celled organisms such as bacteria and archaea, and eukaryotes, which include fungi, plants and animals.
Eukaryotes undergo evolution when mutations are passed down from parent to offspring, known as vertical gene transfer. For prokaryotes, evolution can take place through horizontal gene transfer (HGT for short), where genetic information is directly shared between bacteria. This process allows individual organisms, and even entire species, to quickly gain new genes, including potentially dangerous ones like those that confer antibiotic resistance.
At the University of Delaware, in the lab of Eleftherios “Terry” Papoutsakis, Unidel Eugene du Pont Chair Professor in the College of Engineering’s Department of Chemical and Biomolecular Engineering, the Department of Biological Sciences and the Delaware Biotechnology Institute, researchers recently discovered a new mechanism by which HGT can occur in bacteria. The findings in this study, led by doctoral alumnus Kamil Charubin and doctoral candidate John Hill, expand on the current understanding of evolution and survival strategies for complex microbiomes, with implications for fields ranging from ecology to biotechnology and medicine. The study was published in mBio.
The research for the group’s latest paper began after Charubin observed that two species of bacteria (Clostridium acetobutylicum and C. ljungdahlii) were exchanging nutrients, metabolites and cellular material at high rates when they were in close proximity to one another. They found that the cells were using a mechanism called heterologous cell fusion to transfer materials and wanted to see if bacteria could also transfer genetic information through this mechanism.
“This wasn't just a few proteins but actually encompassed most of the materials in the cytoplasm,” Hill said. “These findings prompted us to determine whether or not genetic material, including plasmids, could be exchanged as well.”
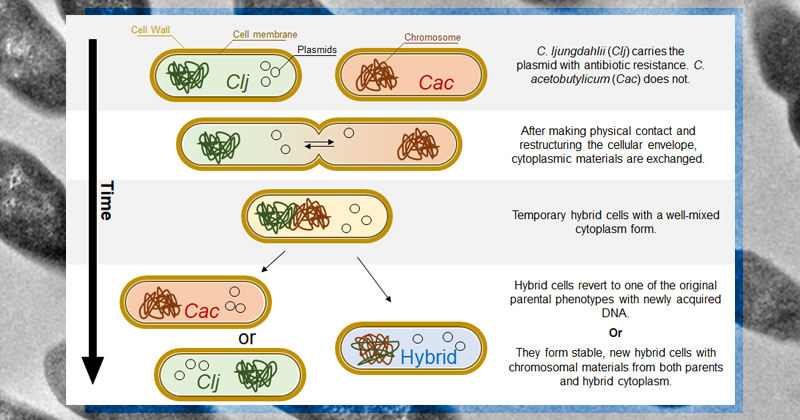
To see if gene transfer was also occurring when the bacterial cells were in close contact, Hill adapted the group’s laboratory techniques so they could track the movement of the bacteria’s genome and plasmids (circular pieces of DNA used by bacteria that are separate from their genome). The researchers also used selective subculturing techniques, isolating C. acetobutylicum cells after they had taken up plasmids from C. ljungdahlii, then confirmed gene transfer using PacBio Single-Molecule Real Time (SMRT) sequencing data.
The results of this research show that the two species of Clostridium can indeed share genetic information via heterologous cell fusion, a mechanism for HGT “that has not been previously contemplated or observed before,” Papoutsakis said. “Through heterologous cell fusion, we found that there is exchange of DNA between the microbes and that the resulting hybrid cells contain large amounts of genomic DNA from both organisms.”
The researchers said that the group’s latest paper provides new insights into the processes and drivers of bacterial evolution.
“We know that microbial life has evolved in naturally occurring communities, and if there is a large number of interspecies interactions, including gene exchange, that would reveal another aspect of microbial evolution,” Hill said. “These results could implicate that microbes are not evolving independently from one another but rather that there exists a multiplicity of evolutionary trajectories within local environments which are motivated by a variety of external pressures, including HGT.”
Papoutsakis added that this study could also have implications in other fields as well, especially if it’s another way that bacteria can confer traits such as antibiotic resistance to one another.
“There's a lot more complexity and interaction between microbes in natural microbiomes, such as those in the environment or the human gut, for example,” Papoutsakis said. “This kind of mechanism for HGT can really have important physiological and medical implications.”
This research was supported by a grant from the Army Research Office (W911NF-19-1-0274), the ARPA-E project under contract (AR0001505) and a U.S. Department of Education Graduate Assistance in Areas of National Need (GAANN) Fellowship to John Hill (P200A210065).
Contact Us
Have a UDaily story idea?
Contact us at ocm@udel.edu
Members of the press
Contact us at 302-831-NEWS or visit the Media Relations website

