Ordered alphabetically by student's last
name
|
|
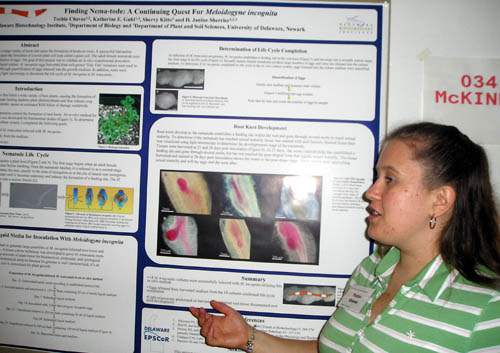
Finding Nema-tode: A Continuing Search For Meloidogyne incognita Toshia Chavez and Janine Sherrier Department of Plant and Soil Science and the Delaware Biotechnology Institute Meloidogyne incognita are plant parasites that infect a large variety of hosts and cause the formation of knots on roots. A successful interaction between the parasite and the host plant is dependent upon the formation of a novel plant cell type called a giant cell. The adult female nematode uses giant cells to acquire necessary nutrients for the production of eggs. The goal of this project was to validate an in vitro experimental procedure developed to infect roots of Medicago truncatula in liquid culture. M. incognita eggs harvested from soil-grown “Tiny Tim” tomatoes were used to infect the roots. The infection process was validated through quantification of eggs released into the growth medium. In addition, roots were harvested at different time points and examined using light microscopy to document the life cycle of M. incognita in M. truncatula. This research has been funded by the NSF EPSCoR grant to Delaware. |
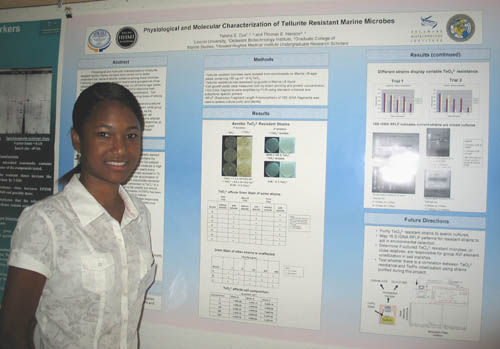
Physiological and Molecular Characterizations of Tellurite Marine Microbes Talisha S. Cox1, 3 and Thomas E. Hanson1, 2 1Delaware Biotechnology Institute, 2Graduate College of Marine Studies, 3Lincoln University Physiological and molecular characterizations of tellurite resistant aerobic marine microbes were carried out to better understand the nature of tellurite resistance among these microbes. Following isolation, tellurite resistant strains were grouped into three categories according to colony morphology on LB marine agar plates. Representatives of each group were analyzed to determine their microbiological, physiological and molecular characteristics. Cell morphology, Gram straining and resistance to varying levels of tellurite were determined. Growth in the presence of tellurite appeared to affect the Gram staining properties of the strains. The three different groups had differing resistance to tellurite: group A -pink colonies, turned out to be the least resistant, while group B-cream colonies, was the most resistant. In all groups, as the concentration of tellurite increased, the protein content per cell increased suggesting that increased tellurite concentrations affected global properties of each strain. Efforts are underway to determine, at the level of 16 S rDNA similarity, whether all members of a given colony morphology group are indeed phylogenetically affiliated. Supported by HHMI. |
|
Brian Demarest1, David J. Hansen2 and Susan
E. White-Hansen3
1Environmental Sciences
Student, Wesley College, Dover, Delaware 2Assistant Professor,
Department of Plant and Soil Sciences, University of Delaware, Newark,
Delaware, and 3Geographic Information Specialist, Research and
Education Center, Georgetown, Delaware
Water is a critical resource
in
|

The Effect of Dissolved Oxygen Concentration on Bacterial Communities in Delaware Inland Bays Darren R. Dolly, David L. Kirchman, Rex R. Malstrom, College of Marine Studies, University of Delaware, Lewes DE 19958 Bacterial communities in the Delaware Inland bays are constantly under strong daily changes of dissolved oxygen concentration during the summer months. However the impact of these changes on bacterial growth rates is unclear. To determine this impact in Deleware Inland Bays, we examined the growth rates of the overall bacterial community, as well as specific phylogenetic subgroups in response to different dissolved oxygen concentrations. Growth rates for specific phylogenetic groups, α-proteobacteria, γ-proteobacteria, Cytophaga-like bacteria, as well as Planctomycetes and sulfur-reducing bacteria were determined by fluorescence in situ hybridization (FISH). We expected that bacterial groups capable of growing under low oxygen conditions would be present in our initial sample. Such groups include the sulfur reducing bacteria (SRB) of the δ-proteobacteria group and anaerobic ammonia oxidizing (annamox) bacteria of the Planctomycetes group. The initial water sample was positive for the presence of annamox but was negative for SRB as determined by results from a polymerase chain reaction assay. Our study suggests that a decrease in dissolved oxygen levels has a negative impact on the overall bacterial community as well as specific subgroups such as α-proteobacteria. However, there was a positive effect on the growth rate of other groups, such as Planctomycetes. The presence of the annamox bacteria in these waters may help increase our understanding of this group as well as its importance in denitrification and its role in the overall nitrogen cycle. Funded by the Department of Energy |
|
Gene Expression
Profiling in Metabolic Tissues of Chickens with Hormonally-manipulated
Lean and Fat Phenotypes
Jessica Hall and Larry A. Cogburn Department of Animal and Food Science Obesity is a major health concern in the United States, where about two thirds of the population is overweight. There are a number of factors that contribute to obesity (genetics, nutrition, hormone balance, etc.). Recently, the chicken has reached model organism status with acquisition of abundant genomic resources (i.e., expressed sequence tags, microarrays, and completed genome sequence). The chicken genome shows high synteny with the chromosomal arrangement of human genes, which makes it an excellent model for studying obesity. The first goal of this project was to use hormone implants (corticosterone, estrogen, and thyroid hormone) to induce lean and fat phenotypes. The second objective was to use microarray analysis to identify major genes that control fat metabolism. The results showed that abdominal fat content of six-week-old chickens was dramatically altered after two weeks of infusion of thyroid hormone or corticosterone. Thus, exogenous corticosterone produced the fat phenotype by stimulating lipogenesis and increasing deposition of body fat. The lean phenotype was induced by exogenous T3, which causes increased metabolic rate and lipolysis. Microarray analysis and quantitative reverse transcriptase-polymerase chain reaction (Taqman®) analysis indicated the differential (induced or suppressed) expression of several candidate genes, which include metabolic enzymes, transcription factors, transport proteins, etc. Additional statistical analysis of the microarray data, coupled with TaqMan® verification is necessary to compile an exhaustive list of the major regulatory genes that control fat metabolism. Genes identified through microarray analysis will be used to functionally map major metabolic pathways responsible for lean and fat phenotypes. Funding is provided by the USDA grant |

Screening a Delaware River Metagenomic Library for Markers Conferring Resistance to Stressor Compounds Nolberto Figueroa Matías, David L. Kirchman, and Thomas E. Hanson Graduate College of Marine Studies, University of Delaware, Lewes, DE 19958 This project seeks to identify specific genes that confer resistance to a particular stressor compound from the genetic pool present in the Delaware River microbial community. The physiological diversity of many microbial communitie remains unknown because of the inability to grow the majority of microbes from environmental samples. The metagenomic approach gives us the opportunity access unknown genes by their cloning and expression in a host organism that can be easily grown in the laboratory. The metagenomic library was constructed in a fosmid vector using DNA isolated from the Delaware River microbial community and was maintained in the Escherichia coli strain EPI 300. Strain EPI 300 carrying the fosmid vector alone was grown in the presence of different stressor compounds, primarily antibiotics and oxidative stress agents, to determine appropriate screening concentrations where growth of the strain was inhibited. Next, pools of metagenome library clones were subjected to stressor compound concentrations determined above to enrich for strains displaying enhanced resistance compared to the vector only control. Enrichments resistant to spectinomycin, ampicillin and sodium tellurite were recovered in this screen. Clonal populations of resistant strains were established from these enrichments and their characteristics examined. Current experiments are underway to verify that the resistance arises from cloned environmental DNA and identify the specific gene conferring resistance. |

Classification of Viral Communities Based on the Fluorescence Intensity of Viral Particles Stained with SYBR Gold Candice M. Johnson1, Danielle M. Winget2, Kurt E. Williamson2, and K. Eric Wommack2 1Lincoln University, 2Department of Plant and Soil Science and Delaware Biotechnology Institute |

The Influence of Marek’s Disease Virus Transcription Regulator Meq on Interleukin-6 Promotor Activity Amanda Kilby, Bruce Kingham, and Carl Schmidt Department of Aninmal and Food Sciences Marek’s Disease (MD) is an immunosuppressive T-cell lymphoma of chickens caused by the herpesvirus MDV. MDV contains a transcription regulator known as Meq, which controls oncogenesis of the disease. Previous experiments have shown, via microarray, that select genes within the host cells (chicken embryonic fibroblasts) are regulated by Meq. Interleukin-6 (IL-6) is a cytokine which modulates the immune system. We hypothesize that IL-6 is regulated by Meq. To test the hypothesis, the IL-6 promotor was cloned and sequenced. We hope to subcloned the promoter to a luciferase reporter plasmid and determine the influence of Meq on the activity of IL-6. Research made possible by funding from the USDA. |

Characterization of 26 Avian Escherichia coli Isolates from the Delmarva Peninsula Suzanne M. King, Cynthia M. Boettger, and John E. Dohms, Department of Animal and Food Sciences Twenty six E. coli isolates from various growers around the Delmarva Peninsula were examined for specific virulence factors and antimicrobial susceptibility. After PCR amplification, 65% of the isolates were positive for the increased serum survival (iss) gene, 42% were positive for the temperature sensitive hemagglutinin (tsh) gene, 46% for the iron uptake chelate in the aerobactin system (IucC), 15% for class I integrase (IntI1), and 19% for TraT, which encodes for an outer membrane protein which may play a role in serum resistance. The isolates were tested for antimicrobial resistance to a fluoroquinolone, ciprofloxacin. Eight percent of the isolates were resistant and 11.5% were intermediate to the ciprofloxacin. Source of Funding: USDA |
|
Andrea J. Laycock* and Kali E. Kniel
Department of Animal and Food Sciences Cryptosporidium parvum and Cyclospora cayetanensis are protozoan parasites that cause severe gastrointestinal illness and are associated with water- and foodborne outbreaks. Both parasites have been linked to consumption of contaminated fresh produce. During the summer of 2004, 100 people became ill after eating raw snow peas contaminated with C. cayetanensis and in the autumn of 2003 several people became ill after drinking ozonated apple cider contaminated with C. parvum. Illness occurs after the ingestion of environmentally stable oocysts, which may contaminate produce through water, soil, or infected food handlers. Transmission is likely due to the small infectious dose of both organisms and the small size of the oocysts (C. parvum 4-5 µm and C. cayetanensis 8-10 µm). In this study the use of ozone as a disinfectant was evaluated on the inactivation of C. parvum in apple cider and Eimeria acervulina on snow peas (surrogate for C. cayetanensis). Ozone was delivered at 0.9 g/L and 2.4 L/min using a portable generator and then recovered oocysts were analyzed in a cell culture infection assay followed by PCR for C. parvum or an in vitro excystation assay for E. acervulina. Ozone was effective at inhibiting the infectivity of C. parvum in a batch system (30 min) for small samples of apple cider and for clarified juices. Ozone treatment for 15 min reduced E. acervulina excystation by >50%. Ozone is currently used in the food industry and has potential for use in the disinfection of fresh produce from protozoa. Work supported by USDA Scholars Grant. |
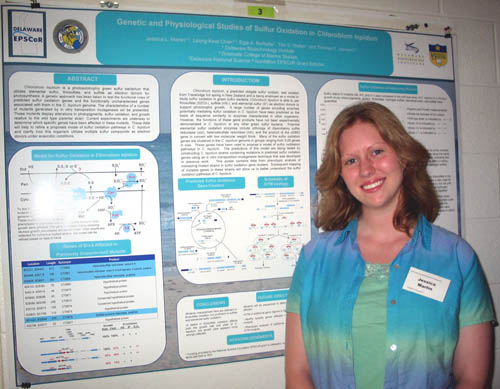
Genetic and Physiological Studies of Sulfur Oxidation in Chlorobium tepidum Jessica L. Martin1,3, Leong-Keat Chan1,2, Egle A. Burbaite1, Tim S. Weber1, Thomas E. Hanson1,2 1Delaware Biotechnology Institute, 2Graduate College of Marine Studies, 3Delaware National Science Foundation EPSCoR Grant Scholar Chlorobium tepidum is a photoautotrophic green sulfur bacterium that utilizes elemental sulfur, thiosulfate, and sulfide as electron donors for photosynthesis. A genetic approach has been taken to test the functional roles of predicted sulfur oxidation genes and the functionally uncharacterized genes associated with them in the C. tepidum genome. The characteristics of a number of mutants generated by in vitro transposition mutagenesis will be presented. These mutants display alterations in photopigments, sulfur oxidation, and growth relative to the wild type parental strain. Current experiments are underway to determine which specific genes have been affected in these mutants. These data will help to refine a proposed model of sulfur oxidation pathways in C. tepidum and clarify how this organism utilizes multiple sulfur compounds as electron donors under anaerobic conditions. |

Validation of Organ-Specific Transcripts from Arabidopsis Flowers Jason Peiffer1, Gervan G. Williams1, Hassan Ghazal1, Hajime Sakai2, Blake Meyers1 1Department of Plant and Soil Science and 2Dupont Crop Genetics, Wilmington DE Massively Parallel Signature Sequencing (MPSS) is a technique invented and commercialized by Solexa, Inc. of Hayward, California. MPSS produces short (17 bp) sequence tags from the 3’ end of an mRNA transcript. Each tag may be traced to a specific gene by comparing the tag sequence to the genomic sequence. The relative abundance of a tag in a given library of transcripts represents the corresponding gene’s expression level. Using the MPSS technology, we have developed five MPSS libraries from the model plant Arabidopsis, corresponding to immature floral tissues of the wild type and the floral homeotic mutants apetala1 (ap1), apetala3 (ap3), agamous (ag) and superman/apetala1 (sup/ap1), all in a Col-0 background. Using a subtractive approach, the flower libraries of the homeotic mutants were compared to the wild type library to identify individual genes specific to the petal, carpel, and stamen whorls. The MPSS data was also analyzed to identify genes expressed in multiple floral organs. We are validating this data by promoter analysis using the Gateway System (Invitrogen), testing cloned promoters of selected whorl-specific genes to determine if they match the MPSS predictions when driving the expression of the reporter genes GUS and GFP. Both these reporter genes are present in the binary (transformation) vector. The cloned promoter-reporter constructs are introduced into Agrobacterium, which is then used to transform wild type Arabidopsis, generating transgenic seeds. In the mature transgenic plants, the expression patterns of the reporter genes will be compared to the expression patterns predicted by the MPSS technology. Supported in part by the USDA. |

Marek’s Disease Meq Oncogene Regulation of the Host Gene Interferon Sean Sheridan1 and Carl J. Schmidt2 1Department of Biology Wesley College, Dover Delaware and 2 Department of Animal and Food Sciences, University of Delaware Marek’s disease is caused by a herpesvirus (Marek’s disease virus, MDV) and yields T-cell lymphomas in chickens. The genome of MDV contains the gene Meq, which encodes a transcription factor that regulates both host and cellular gene expression. Previous work has suggested that the chicken gene for Interferon (IFN) is induced by Meq expression in MDV induced lymphomas. To test this hypothesis, we wished to test directly if the Meq gene product could affect transcription from the IFN promoter. To initiate these experiments, a 2KB fragment containing the chicken IFNgpromoter has been cloned and sequenced. The intent is to subclone this fragment into a luciferase reporter plasmid, and monitor the activity of the IFNg promoter as a function of the level of Meq in co-transfected cells. This project was supported by the NIH NCRR INBRE grant to Delaware, grant number 2P20RR016472-04 |

Trillium Growth in Tissue Culture Jesse Sinanan, David Opalka, Sherry Kitto Department of Plant and Soil Science Trillium, a herbaceous perennial found in the forest understory, has been successfully tissue cultured in the laboratory. It can be grown from either seed or rhizome division. A rhizome is an underground stem that has the potential to generate a flowering shoot. When grown in tissue culture rhizomes generally form a bulbous mass consisting of one large parent rhizome surrounded by many smaller ‘mini’ rhizomes. This project entailed recording growth changes that occurred to five rhizomes of Trillium maculatum in tissue culture. Pictures of each rhizome were taken on intervals of three to four days for approximately two months and a movie compilation of the respective pictures was made for each rhizome. NIH Bridges |