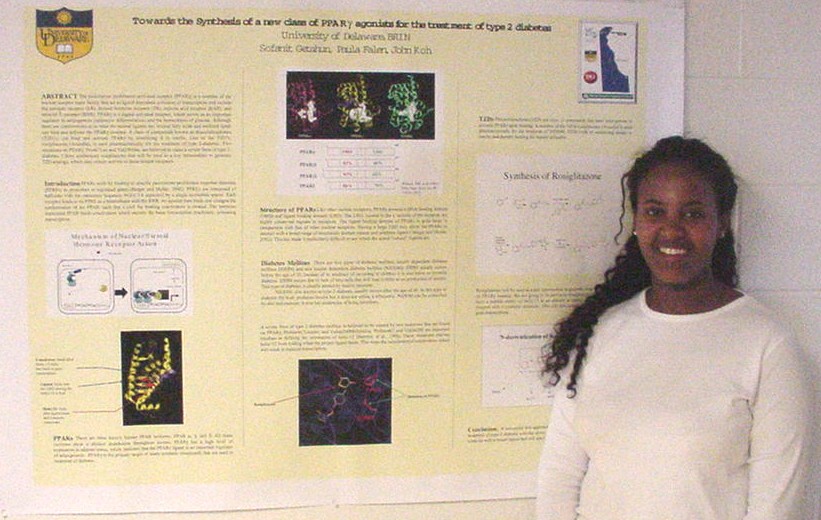
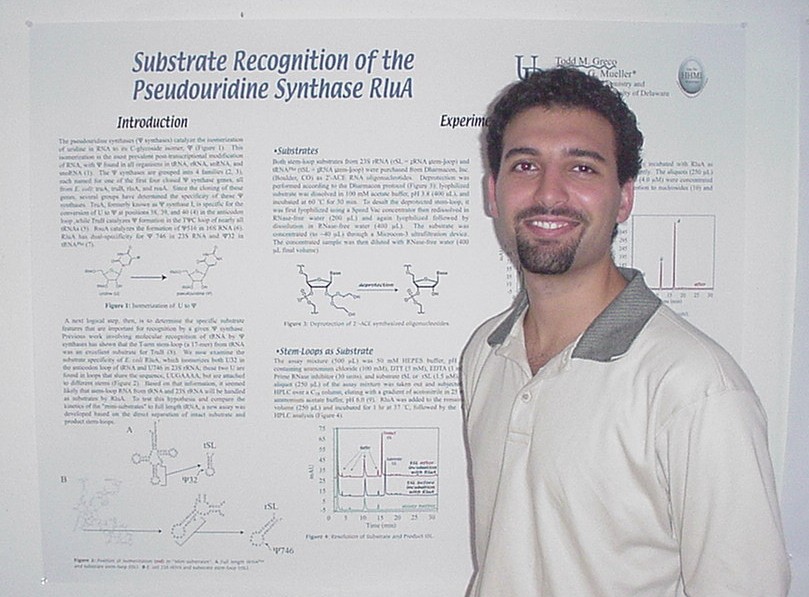
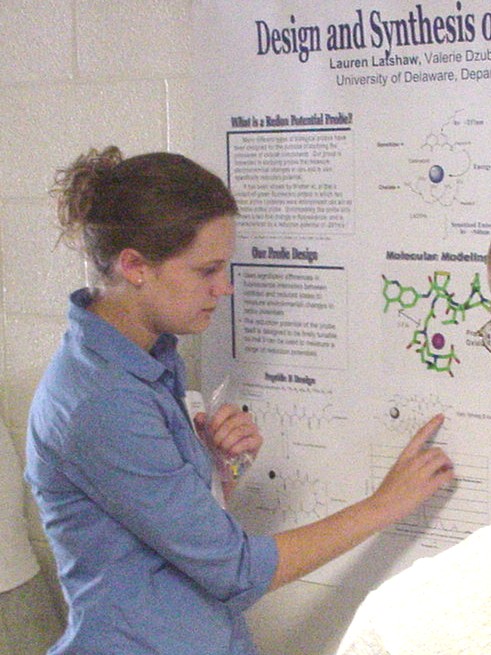
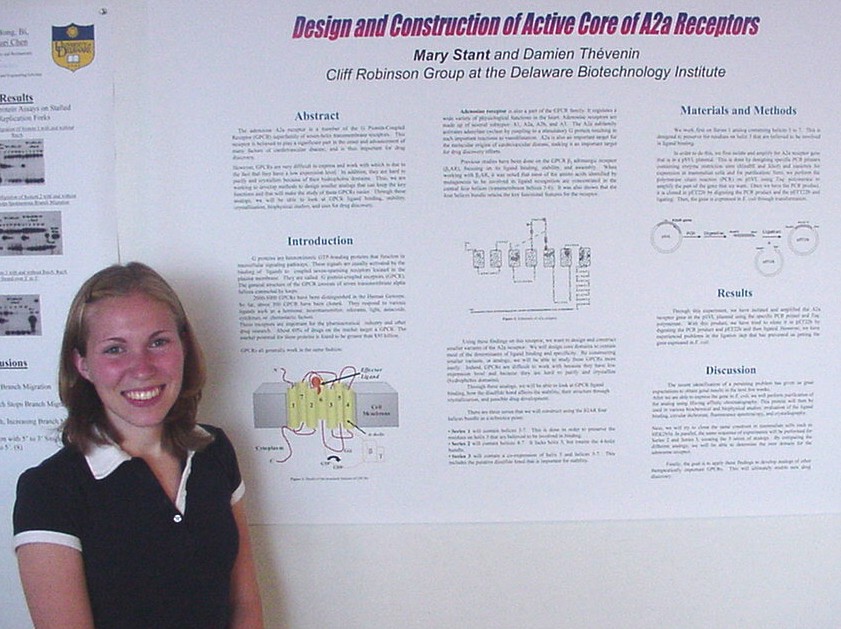
as Novel Substrates of Sulfhydral Oxidase
Steve Brohawn, James Psathas, and Colin Thorpe
Department of Chemistry and Biochemistry
Commercially available trialkyl phosphines such as tris-(2-carboxyethyl)-phosphine (TCEP) and tris-(2-cyanoethyl)-phosphine (TCNP) are used in biochemistry as reductants of disulfide bonds. TCEP, but not TCNP, is a good substrate for chicken egg white sulfhydral oxidase (SOX), a flavoenzyme that catalyzes the oxidation of free thiols to disulfide bonds in a variety of substrates while reducing oxygen to hydrogen peroxide. In an effort to understand these differences in reactivity with the enzyme and possibly to find versatile water soluble phosphines, mono-, di-, and trimethyl ester derivatives of TCEP have been synthesized. Kinetic studies of TCEP, TCNP, and the methyl ester derivatives have been performed on a stopped flow spectrophotometer using DTNB as a model disulfide. The di- and trimethyl ester derivatives have a second order rate constant three times higher than that of TCEP and the monomethyl esters rate constant is close to two and a half times higher than that of TCEP. TCNP was found to be much less reactive than TCEP or its derivatives. SOX activity with TCEP, TCNP, and the trimethyl ester was compared using an oxygen electrode. SOX activity with TCEP gave a Vmax of 1650/min and a Km of 43 mM. The trimethyl ester derivative, like TCNP, was found to have undetectable activity towards SOX. These studies will form the basis for the investigation of the inhibition of SOX with metal ions.